Biology: Biological sequences and genetic code primer.
The members of any biological species are similar in some characteristics but different in others. All human beings share a set of observable characteristics, or traits, that define us as a species: We have a backbone and a spinal cord, we are warm blooded and feed our young with milk from mammary glands, we stand upright and have long legs, relatively little body hair, a large brain, etc. The biological characteristics that define us as a species are inherited (i.e. transmitted from one generation to the next one), but they do not differ from one person to another. Within the human species, however, there is also much variation. Traits such as hair color, eye color, skin color, height, weight, and personality characteristics are largely variable from one person to another. There is also variation in health-related traits, such as predisposition to high blood pressure or diabetes. Not all of these traits are inherited biologically, some are inherited culturally (our native language, for example). Many traits are influenced jointly by biological inheritance and environmental factors. For example, weight is determined in part by inheritance, but also in part by eating habits and level of physical activity.
Fundamental concept of genetics (the study of biologically inherited traits):
| Inherited traits are determined by elements of heredity, called genes, that are transmitted from parents to offspring in reproduction. |
1. DNA - The genetic material.
The chemical substance of the genes, are huge molecules called deoxyribonucleic acid (DNA), that, except for viruses, are part of even huger structures called chromosomes. In eukaryotes (that include all animal and all plants), these chromosomes are located in the cell nucleus. Some details concerning the chromosomes:
|
DNA structure.
Deoxyribonucleic acid (DNA) is a polymer composed of four molecules, called nucleobases, nitrogenous bases, nucleic acid bases, or simply bases. Their names and one-letter abbreviations are adenine (A), cytosine (C), guanine (G), and thymine (T). Two of these bases (adenine and guanine) have a double-ring structure; they are called purines. The other two bases (thymine and cytosine) have a single-ring structure; they are called pyrimidines.
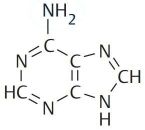 |
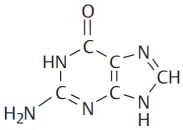 |
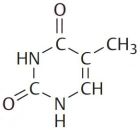 |
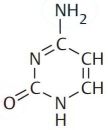 |
|||
| Adenine | Guanine | Thymine | Cytosine |
When a nucleic acid base is N-glycosidically linked to a 2-deoxyribose, it yields a nucleoside (more precisely a deoxyribonucleoside). The derivates of the four DNA bases are called respectively: adenosine, guanosine, thymidine and cytidine (more correctly: 2'-deoxyadenosine, 2'-deoxyguanosine, etc). In the cell, the 5'OH group of the sugar component of the nucleoside is usually esterified with phosphoric acid. This yields the four nucleotides (or nucleosides monophosphate): adenosine monophosphate (adenylic acid), guanosine monophosphate (guanylic acid), thymidine monophosphate (thymidylic acid) and cytidine monophosphate (cytidylic acid). Similar to the nucleosides, a more correct denomination would be 2'-deoxyadenosine-5'-monophosphate, etc. The polymer of these four nucleotides forms a deoxyribonucleic acid (DNA).
In the three-dimensional structure of the DNA molecule proposed in 1953 by Watson and Crick, the molecule consists of two polynucleotide chains twisted around one another to form a double-stranded helix in which adenine and thymine, and guanine and cytosine, are paired in opposite strands. The figure1 below shows the structure of the DNA. At the left: the DNA formula, at the right the DNA double strand.
![DNA structure [Koolman/Roehm, Color Atlas of Biochemistry, 2nd edition, © 2005 Thieme] DNA structure [Koolman/Roehm, Color Atlas of Biochemistry, 2nd edition, © 2005 Thieme]](../pics/dna.jpg) |
Central feature of the DNA structure:
| DNA is composed of two strands (nucleotide chains) held together by the pairing of complementary bases: A with T and G with C. |
This means that on one side, nothing restricts the sequence of bases in a single strand and any sequence could be present along one strand. DNA from different organisms may thus have different base compositions. On the other side, however, because the strands in duplex DNA are complementary, the following, known as Chargaff's rules, is true whatever the base composition is:
|
The amount of adenine equals that of thymine: [A] = [T] The amount of guanine equals that of cytosine: [G] = [C] The amount of purines equals that of pyrimidines: [A] + [G] = [T] + [C] |
Each backbone in a double helix consists of deoxyribose sugars alternating with phosphate groups that link the 3' carbon atom of one sugar to the 5' carbon of the next in line. The two polynucleotide strands of the double helix are oriented in opposite directions in the sense that the bases that are paired are attached to sugars lying above and below the plane of pairing, respectively. The sugars are offset because the phosphate linkages in the backbones run in opposite directions and the strands are said to be antiparallel. This means that each terminus of the double helix possesses one 5'-P group (on one strand) and one 3'-OH group (on the other strand). The figure2 below represents a segment of a DNA molecule showing the antiparallel orientation of the complementary strands.
![DNA antiparallel strands [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers] DNA antiparallel strands [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers]](../pics/dna2.jpg) |
DNA sequences.
We saw above that deoxyribonucleic acids (DNA) are polynucleotides, consisting of deoxyribonucleotide monomers. Even though the tri-dimensional structure of these
molecules is highly complex, it is very easy to represent them in a simple form. In fact, the backbone of the molecule (deoxyribose and phosphate groups) is always
the same, the only variable part of each monomer being the nucleobase. Using the first letter of the base name as a code for this base, we could thus represent the
DNA fragment from the previous figure as something like this:
T G C A T G
| | | | | |
A C G T A C
But, is it really necessary to write down the bases of both strands? No! We saw that the two strands of DNA are always complementary, thus writing down one of them,
we also know the other one. And our DNA fragment could simply be represented by the following DNA sequence:
TGCATG
Why TGCATG and not GTACGT? Or is this the same? It is not the same, because as we saw before, the DNA strands have an orientation (or directionality). When the cell uses the DNA, as for example when transcribing it to RNA, it does so base by base from the 5' end to the 3' end of the molecule. Thus, when DNA is written, it's done so left to right on the page, corresponding to the 5' to 3' orientation of the bases.
Applying this rule to the second strand, the DNA sequence will be written as:
CATGCA
The relationship of the bases of the two strands is described by the expression reverse complement. It's "reverse" because the orientations
are reversed, and "complement" because the bases always pair to their complementary bases, A to T and C to G.
Creating a DNA file, for usage with computer programs is thus really simple: Just open a text editor and enter the sequence of A, T, C and G. Please, note, that normally uppercase letters are used. A reason to use lowercase instead would be to clearly show that the file contains a DNA (or RNA) and not a protein sequence. An issue of doing so would be that not all bioinformatics software accepts lowercase base codes. Also note, that in bioinformatics, sequences are rarely stored as raw text files. In fact, apart from the bases it contains, this tells us nothing about the sequence, in particular there is no indication, what molecule it is, and where it comes from. Over time, biologists and programmers have invented several ways to format sequence data in computer files, in order to create standard representations of sequences, including all kind of information about the sequence. There are lots of such formats; the most widely used are FASTA and GenBank.
FASTA format is basically just lines of sequence data with newlines at the end, so it can easily be printed on
a page or displayed on a computer screen. The length of the lines isn't specified, but for compatibility, it's best to limit them to 80 characters in length. There
is also a FASTA header: one (or several) line(s) at the beginning of the file, and starting with the greater-than (>) sign. The FASTA
header can contain any text (or no text). Typically, a header line contains the name of the DNA or the gene it comes from, often separated by a vertical bar
(|) for additional information about the sequence, the experiment that produced it, or other, non-sequence information of that nature. Most FASTA-aware software insists
that there must be only one header line. The addition of comments (starting with a # character) is not officially supported. This is also the case for multiple sequence
files (several FASTA formatted sequences in the same file). Here how our DNA fragment could look in FASTA format:
>Sample DNA fragment
TGCATG
As not all bases of a sequence are always known, there are also single letter codes for all possible groups of two, three, or four nucleic acids. Here is the complete table of standard IUB/IUPAC nucleobase codes (note, that U = Uracil is a base, present in RNA sequences, corresponding to T in DNA).
|
|
|
DNA replication.
The primary function of any mode of DNA replication is to reproduce the base sequence of the parent molecule. This means that the genetic information, it contains, is precisely inherited by the daughter cells. The specificity of base pairing — adenine with thymine and guanine with cytosine — provides the mechanism used by all genetic replication systems.
| DNA is replicated by unwinding of the two strands of the double helix and building up of a new complementary strand on each of the separated strands of the original double helix. |
Each exposed base has the potential to pair with free nucleotides, present in the cell. Because the DNA structure imposes strict pairing requirements, each exposed base will pair only with its complementary base, A with T and G with C. Thus, each of the two single strands will act as a template to direct the assembly of complementary bases to reform a double helix identical with the original. As each new strand is formed, it is hydrogenbonded to its parental template. As replication proceeds, the parental double helix unwinds and then rewinds again into two new double helices, each of which contains one originally parental strand and one newly formed daughter strand. This is what they call the semiconservative replication of double-stranded DNA. The figure3 below shows the Watson-Crick model of DNA replication.
![Watson-Crick model of DNA replication [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers] Watson-Crick model of DNA replication [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers]](../pics/replication.jpg) |
In reality replication is considerably more complex, because any new DNA strand can be synthesized only in the 5'-to-3' direction. Details concerning the DNA replication:
|
2. mRNA - The protein encoders.
The genetic material DNA contains the information to synthesize the various proteins of the organism, but proteins are not directly synthesized using the DNA information. In fact, there is another polynucleotide that plays the role as an intermediary in the process of decoding genes into polypeptide chains. This polynucleotide actually is a kind of ribonucleic acid (RNA) that, because it passes information, like a messenger, from DNA to protein is referred to as messenger RNA (mRNA).
Beside mRNA there is also so-called functional RNA. They will not be discussed in this tutorial. However, here some details concerning the different kinds of functional RNA:
|
RNA structure.
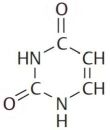
Ribonucleic acid (RNA) is a polymer composed of four molecules, called nucleobases, nitrogenous bases, nucleic acid bases, or simply bases. Their names and one-letter abbreviations are adenine (A), cytosine (C), guanine (G), and uracil (U). Two of these bases (adenine and guanine) have a double-ring structure; they are called purines. The other two bases (uracil and cytosine) have a single-ring structure; they are called pyrimidines.
 |
 |
 |
 |
|||
| Adenine | Guanine | Uracil | Cytosine |
When a nucleic acid base is N-glycosidically linked to a ribose, it yields a nucleoside (more precisely a ribonucleoside. The derivates of the four RNA bases are called respectively: adenosine, guanosine, uridine and cytidine In the cell, the 5'OH group of the sugar component of the nucleoside is usually esterified with phosphoric acid. This yields the four nucleotides (or nucleosides monophosphate): adenosine monophosphate (adenylic acid), guanosine monophosphate (guanylic acid), uridine monophosphate (uridylic acid) and cytidine monophosphate (cytidylic acid). The polymer of these four nucleotides forms a ribonucleic acid (RNA). mRNA molecules are single-stranded and as DNA strands have a 5' and a 3' end. The figure5 below shows an RNA fragment with the two bases guanine and uracil.
![RNA fragment with the two bases guanine and uracil [Koolman/Roehm, Color Atlas of Biochemistry, 2nd edition, © 2005 Thieme] RNA fragment with the two bases guanine and uracil [Koolman/Roehm, Color Atlas of Biochemistry, 2nd edition, © 2005 Thieme]](../pics/rna.jpg) |
RNA sequences.
Ribonucleic acids (RNA) are polynucleotides, consisting of ribonucleotide monomers. Independently from the tri-dimensional structure of these molecules, it is very
easy to represent them in a simple form. In fact, the backbone of the molecule (ribose and phosphate groups) is always the same, the only variable part of each monomer
being the nucleobase. Using the first letter of the base name as a code for this base, we can thus represent an RNA fragment as a sequence of the letters A, U, C or G.
Example:
UGCAUG
Creating a RNA file, for usage with computer programs, may be done by entering the sequence of A, U, C and G in a text editor. Normally uppercase letters are used. A reason to use lowercase instead would be to clearly show that the file contains a RNA (or DNA) and not a protein sequence. An issue of doing so would be that not all bioinformatics software accepts lowercase base codes. Also note, that in bioinformatics, sequences are rarely stored as raw text files. In fact, apart from the bases it contains, this tells us nothing about the sequence, in particular there is no indication, what molecule it is, and where it comes from. Over time, biologists and programmers have invented several ways to format sequence data in computer files, in order to create standard representations of sequences, including all kind of information about the sequence. There are lots of such formats; the most widely used are FASTA and GenBank.
FASTA format has been described, when discussing DNA sequences. Here how our RNA fragment could look in FASTA format:
>Sample RNA fragment
UGCAUG
As not all bases of a sequence are always known, there are also single letter codes for all possible groups of two, three, or four nucleic acids. The complete table of standard IUB/IUPAC nucleobase codes has been given for the DNA bases, when discussing DNA sequences. The codes of this table also apply to RNA, just remembering that T has to be replaced by U. Thus, for example, the pyrimidine code Y means C or T if the sequence is DNA, and it means C or U if the sequence is RNA.
DNA transcription.
DNA represents information, the form at the cellular level is (mostly) constituted by proteins. The biological role of most genes is to carry information specifying the chemical composition of proteins or the regulatory signals that will govern their production by the cell. This information is encoded by the sequence of nucleotides. A typical gene contains the information for one specific protein. The collection of proteins an organism can synthesize, as well as the timing and amount of production of each protein, is an extremely important determinant of the structure and physiology of organisms. As we already said, proteins are not directly synthesized using the DNA information, but there is an intermediary in the process of decoding genes into polypeptide chains: messenger RNA. Please, note that whereas in most cases, the transformation of a DNA sequence results in a mRNA sequence, some DNA sequences are used to create functional RNA.
This brings us to the "central dogma" of molecular genetics:
| DNA codes for RNA and RNA codes for proteins. The step DNA → RNA is called transcription, the step RNA → protein is called translation |
The manner in which genetic information is transferred from DNA to RNA is straightforward. The DNA double helix opens up, and one of the strands is used as a template for the synthesis of a complementary strand of RNA. The process of making an RNA strand from a DNA template is called transcription, and the RNA molecule that is made is called the transcript. The base sequence in the RNA is complementary to that in the DNA template, except that U (which pairs with A) is present in the RNA in place of T in the DNA. Like DNA, an RNA strand also has a polarity, exhibiting a 5' end and a 3' end determined by the orientation of the nucleotides. The 5' end of the RNA transcript is synthesized first and, in the RNA-DNA duplex formed in transcription, the polarity of the RNA strand is opposite to that of the DNA strand. The figure6 below shows the process of transcription as the production of an RNA strand that is complementary in base sequence to a DNA strand, that serves as template.
![DNA transcription [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers] DNA transcription [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers]](../pics/transcription.jpg) |
Key point of DNA transcription:
| During transcription, one of the DNA strands of a gene acts as a template for the synthesis of a complementary RNA molecule. |
The figure7 below summarizes the base-pairing rules between DNA and RNA.
![base-pairing rules between DNA and RNA [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers] base-pairing rules between DNA and RNA [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers]](../pics/transcription2.jpg) |
Writing the transcribed part of the DNA fragment on the transcription figure above as a sequence and transcribing it into a RNA sequence, we get the following:
DNA: CGTGAGATA
RNA: UAUCUCACG
This RNA sequence is obtained by transforming each DNA base into its complement (as the RNA is synthesized as the complement of the DNA template strand), but using U
instead of T, and then reversing the obtained string (in order to write it with its 5' end at the left).
In bioinformatics practice, the transcription is usually done on the direct (not the reverse strand). In this case, there is no need to consider base pairing, nor to
care about orientation. The RNA sequence is identical to the DNA sequence, except that all T have to be replaced by U:
DNA: ATGGAAGTATTTAAAGCGCCACCTATTGGGATATA
RNA: AUGGAAGUAUUUAAAGCGCCACCUAUUGGGAUAUA
Transcribing sequences is thus a very simple procedure. However, the transcription, as it happens in the cells, is a complex process, and there are other related transformations of the mRNA before it can be used to be translated to a protein. All this is beyond the scope of this tutorial. Here just some further details concerning RNA transcription and processing.
|
Reverse transcription.
An unusual polymerase, the reverse transcriptase can use a single-stranded RNA molecule as a template and synthesize a complementary strand of DNA called complementary DNA (cDNA). Like other DNA polymerases, the reverse transcriptase requires a primer. Like any other single-stranded DNA molecule, the single strand of DNA produced from the RNA template can fold back upon itself at the extreme 3' end to form a "hairpin" structure that includes a very short double-stranded region consisting of a few base pairs. The 3' end of the hairpin may serve as a primer for second-strand synthesis. The second strand can be synthesized either by the DNA polymerase or by the reverse transcriptase itself.
Because the introns are absent from the mRNA, the reverse transcription of an mRNA molecule, the resulting full-length cDNA contains an uninterrupted coding sequence for a given protein. If the purpose of forming the recombinant DNA molecule is to identify the coding sequence or to synthesize the gene product in a bacterial cell, then cDNA formed from processed mRNA is the material of choice for cloning.
Example of reverse transcribing an RNA sequence:
RNA: AUUUAAAGCGCCACCUAUUG
DNA: ATTTAAAGCGCCACCTATTG
3. Proteins – The key components of cells.
Proteins are the main macromolecules of an organism. When you look at an organism, what you see is either a protein or something that has been made by a protein. Structural proteins give the cell form and mobility, other proteins form pores in the cell membrane and control the traffic of small molecules into and out of the cell, and still other proteins regulate cellular activities in response to molecular signals from the external environment or from other cells. And all enzymes, i.e. biological catalysts that accelerate biochemical reactions in cells, are proteins. Globally, you can say:
| The products of most genes are specific proteins. The amino acid sequence of the proteins is encoded by the nucleobase sequence in the genes. |
Protein structure.
A protein is a polymer composed of monomers called amino acids. In other words, a protein is a chain of amino acids. Because amino acids were once called peptides, the chain is also referred to as a polypeptide. All amino acids have an amino end (-NH2) and a carboxyl end (-COOH). They also have a side chain, called R (reactive) group. Amino acids general formula:
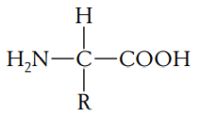 |
There are 20 amino acids known to exist in proteins, each having a different R group that gives the amino acid its unique properties. If you are interested in biochemical details of amino acids, you may want to visit the amino acids page of IMGTeducation.
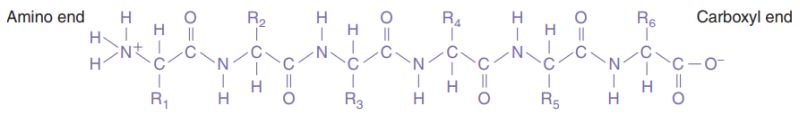
In proteins, the amino acids are linked together by covalent bonds called peptide bonds. A peptide bond is formed by the linkage of the -NH2 end of one amino acid with the -COOH end of another amino acid. Because of the way in which the peptide bond forms, a polypeptide chain always has an amino end and a carboxyl end.
Proteins have a complex structure that has four levels of organization. The linear sequence of the amino acids in a polypeptide chain constitutes the primary structure of the protein. The secondary structure of a protein is the specific shape (α helix, or pleated sheet) taken by the polypeptide chain by folding. The tertiary structure is produced by the folding of the secondary structure. Some proteins have a quaternary structure: such a protein is composed of two or more separate folded polypeptides, also called subunits, joined together by weak bonds. Polypeptide general formula (primary protein structure):
 |
Protein sequences.
Whereas the protein shape and more important its functionality is defined by the tertiary or quaternary structure, the primary structure (amino acid chain) is sufficient to uniquely identify it. This means that a protein may be described by a sequence of letters, where each letter codes for a given amino acid. These 1-letter amino acid codes are for proteins, what the nucleobase codes are for DNA. You should, however, always use uppercase letters. As there is the base code N to designate an unknown base, there is the amino acid code X to designate an unknown amino acid. There are also two codes for uncertain amino acids: B for aspartic acid or asparagine, and Z for glutamic acid or glutamine. These 1-letter codes are fine for being read by computer programs, to make a protein sequence better readable for humans, there are also 3-letter protein codes. Here is the complete table of standard IUB/IUPAC amino acid codes.
|
|
So, we can represent a polypeptide as a sequence of the amino acid codes. Example:
RQNSSTFSAS
and the same with 3 letter-codes:
ArgGlnAsnSerSerThrPheSerAlaSer
As for nucleic acids, protein sequences are rarely stored as raw data, but using a formatted display, that biologists and programmers invented, the primarily reason
for this being to include additional information of the sequence into the file. And as for DNA and RNA, a common format for storing protein sequences is
FASTA (that has been described, when discussing DNA sequences). Simple example of a FASTA formatted polypeptide:
>Some very small sample polypeptide sequence
RQNSSTFSAS
The genetic code.
We have said above that the products of most genes are specific proteins. If genes are segments of DNA and if a DNA strand is just a string of nucleotides, then the sequence of nucleotides must somehow dictate the sequence of amino acids in proteins. There is an obvious analogy with a code: If the nucleotides are the "letters" in a code, then a combination of letters can form "words" representing different amino acids. As we need codes for 20 amino acids, a 2-letter code would not be sufficient, as, for 4 nuclebases, the number of combinations would only be 42 = 16. We'll thus need 3-letter codes, giving 43 = 64 possible combinations. This genetic code is a non-overlapping code, what means that the bases are read sequentially in sets of three and a given base is found in only one code. Example: For the sequence UAUCUCACG, and supposing that translation starts with the first base, the coding triplets are UAU, CUC and ACG, but not: AUC, UCU, UCA, and CAC. Genetic code definition:
| Each sequence of three adjacent bases in mRNA is a codon that specifies a particular amino acid (or chain termination). The genetic code is the list of all codons and the amino acid that each one encodes. |
The figure8 below shows the genetics code table. The chart is for DNA (using DNA codons) rather than for RNA (that is used for the translation). The codons are obtained by concatenating the letters of the inner, the middle and the outer circle. Amino acids are given by their conventional abbreviation in one-letter and three-letter format. Note that the codon AUG (which codes for methionine) is usually used for initiation (start codon; cf. next section) and that some codons don't code for an amino acid but to terminate the translation (stop codons; cf. next section). The codons are to be considered with the 5' base on the left and the 3' base on the right.
![Genetic code chart for DNA [http://www.geneinfinity.org/sp/sp_gencode.html] Genetic code chart for DNA [http://www.geneinfinity.org/sp/sp_gencode.html]](../pics/gencode.jpg) |
If we take the sequence UAUCUCACG, using this table, we can decode the codons into amino acids: UAU → Tyr; CUC → Leu; ACG → Thr.
RNA translation.
The synthesis of the proteins by chaining up amino acids as coded by the base sequence of the corresponding gene is called translation. In
the simplest case (not actually what happens in the cell), we can consider that a given DNA fragment is transcribed base by base from the beginning to the end
(this transcription being nothing more than replacing all T by U) and that then, the resultant RNA sequence is translated from the beginning to the end into a
polypeptide (this translation being creating a protein sequence with one given amino acid for each RNA codon). Example:
DNA: ATGGAAGTATTTAAAGCGCCACCTATTGGGATATA
RNA: AUGGAAGUAUUUAAAGCGCCACCUAUUGGGAUAUA
protein: MEVFKAPPIGI
Note that in this example, the last two bases ("incomplete codon") have been ignored when doing the translation. This puts up the question if, instead of ignoring bases at the end of the sequence, we couldn't or shouldn't ignore them at its beginning. The fact is, that often we don't know where the translation actually starts. Only about 1-1.5% of human DNA is in genes, which are the parts of DNA used for the translation into proteins. Furthermore, genes very often occur in pieces that are spliced together during the transcription/translation process. Since the codons are three bases long, the translation happens in three "frames", for instance starting at the first, the second, or the third base (the fourth would be the same as starting from the first). Each of these 3 starting places (biologists call this 3 different reading frames) gives a different series of codons, and, as a result, a different series of amino acids.
Also, transcription and translation can happen on either strand of the DNA; i.e. either the DNA sequence, or its reverse complement, might contain DNA code that is actually translated. The reverse complement can also be read in any one of three frames. So a total of six reading frames have to be considered when looking for coding regions, that are part of the DNA that encodes proteins.
Reconsider the example above, doing this time a 6 reading frames translation:
Strand 1:
DNA: ATGGAAGTATTTAAAGCGCCACCTATTGGGATATA
RNA: AUGGAAGUAUUUAAAGCGCCACCUAUUGGGAUAUA
RF 1: MEVFKAPPIGI
RF 2: WKYLKRHLLGY
RF 3: GSI*SATYWDI
Strand 2:
DNA: TATATCCCAATAGGTGGCGCTTTAAATACTTCCAT
RNA: UAUAUCCCAAUAGGUGGCGCUUUAAAUACUUCCAU
RF 1: YIPIGGALNTS
RF 2: ISQ*VAL*ILP
RF 3: YPNRWRFKYFH
In reality, a mRNA sequence is not translated from beginning to end. Translation is normally initiated at the start codon AUG (that also codes for the amino acid methionine) and it is terminated when one of the three stop codons UAA, UAG, UGA is encountered (thus the codon preceding UAA, UAG, or UGA will be the last that is actually translated).
Example of translation with start and stop codons:
DNA: AGGGAAGTATTTATGGCGCCACCTATTGGGTAGATATA
RNA: AGGGAAGUAUUUAUGGCGCCACCUAUUGGGUAGAUAUA
protein: MAPPIG
In this case, translation is done from start codon AUG at position 13 to stop codon UAG at position 31. Translated RNA length = 18 nucleotides, polypeptide length = 6
amino acids.
As you can imagine, the translation of a mRNA sequence to a polypeptide sequence, as it happens within the cell, is something really complex. Here some details concerning the translation process:
|
Redundancy of the genetic code.
With 3 codons coding for a stop signal, there are 43 - 3 = 61 codons that specify amino acids. In many cases several codons direct the insertion of the same amino acid into a polypeptide chain. The genetic code is said to be redundant (degenerate). In the actual genetic code, all amino acids except tryptophan and methionine are specified by more than one codon. The redundancy is not random. With the exception of serine, leucine, and arginine, all codons that correspond to the same amino acid (they are called synonymous codons) usually differ only in the third base. For example, GGU, GGC, GGA, and GGG all code for glycine. Moreover, in all cases in which two codons code for the same amino acid, the third base is either A or G (both purines) or T or C (both pyrimidines).
Substitution point mutations (mutations concerning one single nucleobase, that is replaced by another one, thus changing the base sequence of a gene), may differ in their consequences. A missens mutation results in the replacement of one amino acid by another. For example, the change from GAG to GTG in the DNA of the gene for β-globin results in the replacement of glutamic acid by valine in the sickle-cell hemoglobin molecule. In contrast, a silent mutation is one that does not change the amino acid sequence. Silent mutations often result from changes in the third codon position. For example, a mutation that changes an AAA codon into an AAG codon is silent because both codons specify lysine. Another class of mutations consists of changes that convert a codon that specifies an amino acid into a stop codon. Such a mutation is called a nonsense mutation, and it results in premature termination of the polypeptide chain. An example is found in the β-globin gene, in which a mutation from AAG to TAG in the seventeenth codon results in a truncated polypeptide with only 16 amino acids in length. This mutation is one of several types associated with the disease β-thalassemia. The contrary is also possible, a stop codon being changed into a coding one; for example: A mutation from TAG (stop codon) to TAC or TAT (tyrosine) results in a chain elongation (a mutation from UAG to UAA being silent). In this case, the protein can loose or not all its functionality.
Annexes.
External references.
The text of this tutorial is based on these two genetics books:
Anthony J. F. Griffiths et. al., An Introduction to Genetic Analysis, 8th Edition, © 2005, W.H. Freeman, New York
Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers
The figures shown in the tutorial have been taken from:
[1] Koolman/Roehm, Color Atlas of Biochemistry, 2nd edition, © 2005 Thieme
[2] Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers
[3] Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers
[4] Anthony J. F. Griffiths et. al., An Introduction to Genetic Analysis, 8th Edition, © 2005, W.H. Freeman, New York
[5] Koolman/Roehm, Color Atlas of Biochemistry, 2nd edition, © 2005 Thieme
[6] Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers
[7] Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers
[8] http://www.geneinfinity.org/sp/sp_gencode.html
[9] Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers
Related computer applications available on my website.
- DNA basics: Molecular genetics desktop application primarily concerning the genetic code (transcription, reverse transcription, translation considering or not the start and stop codons), but also doing some elementary sequence analysis (counts, molecular weight calculation). Lazarus/Free Pascal source code included.
- Point mutations: Simple desktop application concerning point mutations (transitions, transversions, indel mutations), that may be used for mutations analysis or as mutations exercise generator. Lazarus/Free Pascal source code included.
- DNA molecular weight calculator: Online application, that may be used to calculate the molecular weight of DNA sequences entered manually or uploaded from a file. Perl source code included.
If you find this tutorial helpful, please, support me and this website by signing my guestbook.
![DNA replication at the growing fork [Anthony J. F. Griffiths et. al., An Introduction to Genetic Analysis, 8th Edition, © 2005, W.H. Freeman, New York] DNA replication at the growing fork [Anthony J. F. Griffiths et. al., An Introduction to Genetic Analysis, 8th Edition, © 2005, W.H. Freeman, New York]](../pics/replication2.jpg)
![RNA translation: First steps in elongation [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers] RNA translation: First steps in elongation [Hartl/Jones, Genetics: Principles and Analysis, 4th Edition, © 1998 Jones and Bartlett Publishers]](../pics/translation.jpg)